
Dr. Udupi A. Ramagopal
Associate Professor & Dean Academics
Biological Science Division
Email: udupi.ramagopal[at]poornaprajna[dot]org
Structural Biology of enzymes and immune receptors
A biological cell is a highly complex and organized soup of proteins, DNA, RNA, lipids and many other complex molecules that work in synergy. Although, the information necessary for the energy metabolism, defense,sensing the external world, and all other essential processes for the survival are hidden inside the genome of the cell, the real workhorse molecules are the proteins. These proteins can synthesize highly complex molecules essential for everyday life, act as framework for the cell shape and integrity, sense external molecules and also the possible threat from the outside world and so on. Over the course of evolution, nature has learnt all the tricks to create these nano-machines (proteins and their complexes), which are polymers made of just twenty different amino acids. We are interested in understanding these magical-machines, one at a time or when they are talking to each other, using biophysical and biochemical techniques. Since, the visible light has its own limitation to see such objects at atomic details, we use X-rays (X-ray diffraction from ordered 3D-array of these molecules, called crystals) to visualize such molecules. For example, we are interested in structure based functional characterization of key bacterial enzymes responsible for synthesis of essential small molecules for their survival. We are also interested in the molecules on the surface of the immune cells, so called “immune receptors” that recognize the external enemy like bacteria or our own cancer cells and play a critical role in the clearance of such threats.
Protein structure and their importance in biology.
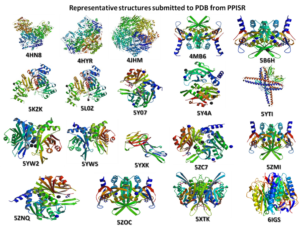
Proteins are the key molecules life and are responsible for most, if not all the processes that take place in any living organism. For example, when we breathe, walk or even sleep, there are different proteins working in an optimal concentration and speed depending upon the need. Most drugs we consume are the modulators of the function of these proteins. Understanding the structure of proteins is essential for understanding the basic biology as well as to design drugs against these proteins. Our lab tries to see these molecular machines in atomic details and also attempts to design small molecular drugs against these proteins. We also try to tinker them for therapeutic applications. The picture below gives the cartoon some of the protein structures from PPISR determined by our group.
- Chatterjee S., Kundapura SV, Basak AJ, Das AK, Samanta D, Ramagopal UA*. High-resolution crystal structure of LpqH, an immunomodulatory surface lipoprotein of Mycobacterium tuberculosis reveals a distinct fold and a conserved cleft on its surface, 2022, International Journal of Biological Macromolecules, 210, PP 494-503. Impact factor 6.95.
- Lankipalli S, , Mahadeva Swamy HS, Selvam D, Samanta D, Nair D, and and Ramagopal UA*. Cryptic association of B7-2 molecules and its implication for clustering. 2021, Protein Science, 30(9), 1958-1973, Impact factor 6.73.
- Lankipalli S, Ramagopal UA*. Structural perspective on the design of decoy immune modulators, 2021, Pharmacological Research, 170, 105735. Impact factor 7.65.
- Liu W, Chou T-F, Garrett-Thomson SC, Seo G, Fedorov E, Ramagopal UA, Bonanno JB, Kakugawa K, Cheroutre H, Kronenberg M, Almo SC. HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD16, 2021, Journal of Experimental Medicine, 218 (12): e20211112. Impact factor 14.3.
- Kundapura S. and Ramagopal UA*. CC’ loop of IgV domains in Immune checkpoint inhibitors play pivotal role in affinity modulation, (2019), Scientific Reports, Dec 16, 9(1):19191. DOI 10.1038/s41598-019-54623-y. Impact Factor 4.1.
- Liu W, Garrett SC, Fedorov EV, Ramagopal UA, Garforth SJ, Bonanno JB, Almo SC, Structural Basis of CD160 HVEM Recognition, (2019), Structure, 6;27(8):1286-1295.
- Pavithra GC and Ramagopal UA*, Crystal structures of APRT from Francisella tularensis: An N-HLN hydrogen bond imparts adenine specificity in adenine phosporibosyltransferases, (2018), FEBS J, 285(12):2306-2318. Impact factor 4.74.
- Ghosh A, Ramagopal UA, Bonanno JB, Brenowitz MD, Almo SC. Structures of the L27 domain of Disc Large homolog 1 protein illustrate a self-assembly module, (2018), Biochemistry, 57 (8), pp 1293–1305. Impact factor 3.0.
- Raghurama P. Hegde, Pavithra G. C., Debayan Dey, Ramakumar S, Ramagopal UA*, Can the propensity of protein crystallization be increased by using systematic screening with metals?, (2017), Protein Science, 26(9), 1704-1713. Impact factor 6.75.
- Ramagopal UA, Liu W, Garrett S, Yan Q, Srinivasan M, Wong S, Bell A, Mankikar S, Vangipuram R, Deshpande S, Korman A, Almo, SC., Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor Ipilimumab, (2017), Proc. Nat. Acad. Sci. (USA), 114(21), E4223-E4232. (This article is highligted in “In this Issue” section of PNAS and the article was edited by Prof. James P. Allison, Nobel Laureate 2018). Comments and media coverage: http://www.deccanherald.com/content/623201/bodys-battle-against-cancer.html and https://medicalxpress.com/news/2017-05-cancer-immunotherapy-drugs-x-ray-crystallography.html and https://immuno-oncologynews.com/2017/06/06/insights-into-yervoy-and-target-molecule-may-improve-immunotherapies/ and https://www.natureindex.com/article/10.1073/pnas.1617941114 and http://cancerimmunolres.aacrjournals.org/content/5/7/515
- Raghurama P. Hegde, Fedorov, A.A., Sauder, J.M., Burley, S.K., Almo, S.C. & Ramagopal UA*, The hidden treasure in your data: phasing with unexpected weak anomalous scatterers from routine data sets, (2017), Acta Cryst F, 73, 184-195.
- Samanta D, Guo H, Rubinstein R, Ramagopal UA*, Almo SC, Biochemical and Structural studies reveal a canonical mode of molecular recognition between immune receptor TIGIT and nectin-2, (2017), Molecular Immunology 81, 151-159.
- Lázár-Molnár E, Scandiuzzi L, Basu I, Quinn T, Sylvestre E, Palmieri E, Ramagopal UA, Nathenson SG, Guha C, Almo SC. Structure-guided development of a high-affinity human Programmed Cell Death-1: Implications for tumor immunotherapy, (2017), EBioMedicine, 17:30-44.
- Bhowmick T,Ghosh S, Ramagopal UA, Dey D, Ramakumar S, Nagaraja S. 2014, Structure based inhibition provides an insight into the HU mediated regulation in Mycobacterium tuberculosis. Nature Communications, 5, 4124. JIF =10.7. Comments and media coverage:http://bangaloremirror.indiatimes.com/bangalore/others/iisc-discovery-DNA-TB-Bacteria-indian-institute-of-science-tapasya-mitra-mazumder/articleshow/36562763.cms and https://indiabioscience.org/news/2014/histone-like-proteins-in-bacteria-could-be-potential-drug-targets-against-tuberculosis and www.iisc.ac.in/wp-content/uploads/2016/03/Nagaraja_Ramakumar_Final.pdf and http://www.thehindu.com/sci-tech/health/targeting-a-chink-in-tb-bacterias-armour/article6127017.ece
Read More
Sponsored projects
● Ramalingaswami Fellowship titled “Co-stimulatory molecules: Biology and therapeutic intervention”, Department of Biotechnology (DBT), New Delhi, India.
● Design of modified B7-1 (CD-80) and B7-2 (CD86) molecules to create potential reagents for cancer and auto-immune disorders”, Vision Group on Science and Technology (VGST), Karnataka (Ms. Swetha, L.).
● Structural and evolutionary investigations on antibiotic resistance conferring rRNA methyltransferases for designing novel strategies of drug development, Department of Science and Technology, India.
● Bristol Myers sqibb (BMS) scholarship Grant to Udupi A. Ramagopal, titled “Structure based rational design of Pd1 mutants to create lead molecule for cancer immunotheraphy.
● X-ray data collection and travel grant to synchrotron sources at (1) SOLEIL, France, ESRF, France (2 proposal approved), XRD2, Trieste, Italy (2 proposal approved) and X29 USA.
In-house projects
● Structural Studies of purine Phosphoribosyltransferases from Pathogenic Bacteria (Ms. Pavithra G. C.).
● Structural Study of proteins from the enolase superfamily (Dr. R. P. Hegde - Finished)
● Testing the limits of macromolecular crystallographic phasing (Dr. R. P. Hegde- FInished)
● Associate Professor & Dean Academics, 2019 - Current, Poornaprajna Institute of Scientific Research, Bangalore, India.
● Associate Professor (Ramalingaswami Fellow - DBT), 2014–2019, Poornaprajna Institute of Scientific Research, Bangalore, India.
● Assistant Professor (Ramalingaswami Fellow – DBT 2011-2016), 2011 – 2013, Poornaprajna Institute of Scientific Research, Bangalore, India.
● Visiting Faculty, 2011 – Present: Albert Einstein College Of Medicine, New York, USA. http://www.einstein.yu.edu/home/faculty/profile.asp?id=9276
● 2009-2011: Instructor (Faculty), Albert Einstein College Of Medicine, New York, USA.
● 2005-2009: Associate of Biochemistry (Faculty), Albert Einstein College Of Medicine, New York, USA.
● 2003-2005: Senior Research Associate, Department of Biochemistry, Albert Einstein College Of Medicine, New York, USA.
● 2001-2003: Visiting Fellow, National Institute of Health, USA.
● 2001: PhD, Department of Physics, Indian Institute of Science, Bangalore, India.
- Ramalingswami fellow, DBT, India (2011 - 2016).
- Best thesis “Kumari L. A. Meera Award and a Gold Medal”, 2001, IISc, India.
- Visiting Fellow (2001 – 2003, NIH, USA).
- Visiting Faculty (Albert Einstein College of Medicine, 2011 – current).
- Invited Instructor (2003-2010) at RapiData, a comprehensive course offered at Brookhaven National Laboratory for budding crystallographers around the world (http://www.bnl.gov/rapidata/).
- Coauthored publications with Prof. James Allison, Nobel Laureate in Physiology and Medicine 2018.
- Proposal reviewer: Macromolecular Crystallography, APS, Argonne National Laboratory.
- Served in the "User Executive Committee 2002-2003" of National Synchrotron Light Source, Brookhaven National Laboratory, USA.
- Jeffery Award (poster award - IUCr 2002, co-author).
- Contributed > 250 protein structures to World Wide Protein Data Bank (wwPDB).
- Referee for various International Journals such as Acta Crystallographica Section-D, Protein Science, BioChem Journal, Nature Scientific Reports, VirusDisease and so on.
- Doctoral Advisory committee member for a few students registered under Manipal University.
- Scientific Advisor “Genelon Life Science Ltd.”, Yelahanka, Bangalore.

Ms. Swetha L. (Graduate Student)
I did my B.Sc.Ed at the Regional Institute of Education, Mysore and masters in Zoology at Sri Venkateswara University, Tirupati where I am a recipient of Gold medal. I am also a recipient of CSIR-JRF (2012) and qualified Graduate Aptitude Test in Engineering (GATE-2012) and Andrapradesh State Elligibility (AP-SET-2012). I joined the structural biology group at PPISR in 2013 and currently I am a senior research fellow here.
Boosting the body’s immune system to fight against an ailment is often referred to as immunotherapy. Modulation of T-cell co-stimulatory/inhibitory pathways has been proven to be one of the main immunotherapeutic approach in the treatment of cancer. CD28/CTLA-4: B7-1/B7-2 family of molecules being one of the key proteins in the T-cell co-stimulatory/ inhibitory pathway are most explored targets for immune check-point inhibitors. This project aims to modify human B7-2 protein, aided by the CD28 and CTLA-4 structure analysis, to make it a better immune-modulator than the existing antibody-based drugs.

The T-lymphocyte cells (T-cells) are the sentinels of the cell-mediated immune response. They play a vital role in eliminating specific challenges to our body such as tumor cells, infectious agents and the like. However, their actions are very nuanced and complex. One such example is of the immune checkpoint receptors present on T-cells and few other immune related cells. These immune checkpoint receptors modulate the extent of T-cell activity. These receptors are responsible for maintaining a delicate balance between the autoimmunity and diseased state.
One such immune checkpoint receptor is of particular interest namely, programmed cell death protein-1 (PD-1). This receptor-ligand pathway is responsible for dampening the T-cell immune response. The tumor cells exploit this pathway to escape immune surveillance by the T-cells. A myriad of antibodies have been used to remove this advantage enjoyed by the tumor cells, to various degrees of success. However, issues of toxicity and unassailable cost of these antibodies demand a cheaper and safer alternative. An attempt is being made to address these issues by rationally and minimally modifying the receptors and employing them as an alternative to antibodies- Mr. Akshaya Bhat (Graduate Student)
- Ms. Salima Parveen (Graduate Student)
PAST MEMBERS (Alumni)

Ms. Pavithra G. C. (Post-doctoral Fellow, NCBS, Bengaluru)
Dr. Pavithra obtained M. Sc. in Microbiology from Bangalore University. Towards her PhD, she worked on the enzyme involved in salvage of purines. Purine phosphoribosyltransferases are a glycosyltransferase that catalyse the reaction between purine and PRPP to synthesize respective purine monophosphate (AMP/GMP) with the release of pyrophosphate (PP). In many pathogenic bacteria that lack the de novo pathway, salvage pathway is the only way to produce purine nucleotides which are essential components of DNA/RNA and also precursors for biologically important molecules such as ATP/GTP so on. Hence, Dr. Pavithra’s thesis work concentrated on structural and functional characterization of PRTases from pathogenic bacteria, which lack the de novo purine synthesis. Currently, she working as a post-doctoral fellow at,
Dr. Sachi Gosavi’s Lab, NCBS, Bengaluru.

Dr. Raghurama P. Hegde
(Beamline Scientist, Elettra Synchrotron Beamline, Italy)
Raghurama Hegde obtained his M.Sc. in Physics from Christ College (now Christ University), Bangalore University. He is a recipient of Prof. S. P. Bondade Memorial Lecture Contest held by the Indian Physics Association (second prize), qualified in the Graduate Aptitude Test in Engineering (GATE) and also recipient of a Junior Research Fellowship and Senior Research Fellowship of the Council of Scientific and Industrial Research (CSIR). He obtained his Ph.D. in Biocrystallography from the Department of Physics, Indian Institute of Science, Bengaluru. Following his PhD, he has moved to Protein Chemistry Laboratory, Department of Biological Sciences, National University of Singapore, Singapore as a Research Fellow.
Dr. Hegde, joined the Division of Biological Sciences, PPISR as a Research Associate. He has worked on two projects: (i) Structural Study of proteins from the enolase superfamily. (ii) Testing the limits of macromolecular crystallographic phasing. In the first project he has determined the crystal structures of three proteins from the enolase superfamily involved in bacterial metabolism. The second project is about importance of looking at all the X-ray diffraction data for weak and serendipitous anomalous signal from unexpected anomalous scatterers in the crystals lattice or from native sulphurs. Later he worked on the structural studies of a putative rRNA methyltranferase from Sinorhizobium meliloti. Dr. Raghu moved to Prof. Silvia Onesti’s Laboratory at Elletra, Trieste, Italy and now he is taking the beamline scientist position at XRD2 beamline, Elettra, Italy.

Dr. Debayan Dey PostDoctoral Fellow, Emory University, USA.
Dr. Debayan received his B.Sc in Physics from Presidency Collage, Kolkata and M.Sc in Biophysics and Bioinformatics from Calcutta University securing a silver medal. Following which he was a graduate student at Dept. of Physics, Indian Institute of Science (IISc). In 2017, he was awarded with a PhD degree from IISc, an internationally recognized and prestigious institute in India. Dr. Debayan is a recipient of meritorious BSR fellowship, awarded by UGC, obtained, first-prize at the ABLE entrepreneurship competition 2010. He joined Dr. Ramagopal's Lab at PPISR in August 2016 and received the “Research Associate Fellowship '' of Department of Biotechnology (DBT) and DST-SERB National Post-Doctoral fellowship. During his tenure at PPISR, he has worked towards understanding the molecular mechanism of antibiotic resistance conferring rRNA methyltransferases’ specificity towards different rRNA targets and develop drugs to break the resistance. Currently, he is a CFF Postdoctoral Fellow at Emory University, USA
MS Thesis:
- Mr. Saurav Kampa (Geetham University; degree awarded, currently at TIGS, Bengaluru).
-
Mr. Deepak K.J (Bangalore University; degree awarded, currently at Indian Immunologics, India)
-
Ms. Kalpana Chaco (Amrita University. ongoing)
-
Ms. Shythya Murali (Amrita University. ongoing)
-
Also, co-mentored several students and post-doctoral fellows on the structural-immunology projects at Albert Einstein College of Medicine

